Serviços Personalizados
Journal
Artigo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Acessos
Acessos
Links relacionados
-
 Similares em
SciELO
Similares em
SciELO
Compartilhar
Portugaliae Electrochimica Acta
versão impressa ISSN 0872-1904
Port. Electrochim. Acta vol.29 no.5 Coimbra set. 2011
https://doi.org/10.4152/pea.201105361
An Electrochemical Biosensor for Acrylamide Determination: Merits and Limitations
Nelson A.F. Silva1,*, Manuel J. Matos1, Amin Karmali1, Maria M. Rocha2
1 CIEQB-ISEL - Chemical Engineering and Biotechnology Research Center, Portugal
2 DQB-FCUL - Chemistry and Biochemistry Department, Portugal
doi: 10.4152/pea.201105361
Abstract
The present work reports the results concerning the development and implementation of the first electrochemical biosensor for acrylamide determination, based on a direct biochemical interaction between the analyte and intact bacterial cells, with intracellular enzymatic activity. The biological recognition element consisted of whole cells of Pseudomonas aeruginosa containing intracellular amidase activity, which catalyses the hydrolysis of acrylamide producing ammonium ion (NH4+) and acrylic acid. The transduction process was accomplished by means of an ammonium ion selective electrode. Whole cells were firstly immobilized on single discs of polymeric membranes, such as polyethersulphone, nylon and polycarbonate, which were, then, attached to the surface of the selective electrode. However, it was observed a significant loss of cells each time the biosensor was used, namely at the beginning of the assay, when the membranes were attached to the ammonium electrode, and after the assay, when removed for storage purposes. This evidence determined a premature decrease in the biosensor's stability. Instead of using single membrane discs, a ''sandwich'' design, with two membrane discs was considered. This way the cells remain contained between the membranes, never contacting the electrode's surface, preventing their premature loss. Consequently, the activity of the biosensor could be maintained for longer periods of time. The analytical performance of the biosensor was evaluated. The best results were obtained when polyethersulphone double membranes were used. A typical response of 120 mV (after 6 min reaction time), a Nernstian slope of 48 mV/decade, a limit of detection of 6.31×10-4 M and a half-life time of 27 days, are examples of some figures of merit observed for this biosensor.
Keywords: biosensor, acrylamide, amidase, whole cells, Pseudomonas aeruginosa, polymeric membranes
Introduction
Acrylamide is a chemical compound with potentially hazardous effects on environment and human health such as muscular weakness, skin and mucous irritation, nausea, numbness and nervous system damage. Long term exposure to acrylamide may also result in the development of several types of cancer [1-3]. Environmental effects include death of animals and low growing rate in plants. Accumulation in groundwater may also occur, as well as persistence in aquatic environments [4, 5].
Acrylamide is also present in some industrial processes, for example in wastepaper recycling, synthesis of dyes, as a soil conditioning agents, as a copolymer for contact lenses or in textile industry. It is also used as an additive in waste and drinking water treatment (where relative high levels of acrylamide can be found) [6, 7].
On the other hand, it was recently discovered that acrylamide is formed when certain types of food, namely those with high contents of carbohydrates and low in protein, are cooked or processed at high temperatures. Although acrylamide formation process in these conditions is poorly understood, it is considered that reducing sugars and asparagines are the main responsible for the high levels of this carcinogenic amide in such foods [8, 9].
In this regard it is essential to develop methods in order to determine, reduce and control the amount of acrylamide present in the environment, namely waste and natural waters, as well as in food, human and animal organisms.
Traditionally the determination of acrylamide is performed by chromatography, namely Liquid Chromatography with Mass Spectrometry detection (LC/MS) [10], which is a time consuming and expensive process. Therefore, the development of a biosensor for acrylamide may result in a significant alternative towards traditional techniques, where several advantages like analyte specificity, easy operation and low cost equipment can be stressed out.
In fact, one of the main advantages of using whole cells of Pseudomonas aeruginosa, as the biological recognition element, instead of cell-free extract containing enzyme or purified enzymes, is the significant decrease in time consuming and costs. Some procedures like cell sonication or enzyme purification are overcome.
This work reports the development of an electrochemical biosensor for acrylamide determination, using an ammonium ion selective electrode (ISE) [11, 12] as an electrochemical transductor. The biological recognition element, or biocomponent, consisted of whole cells of Pseudomonas aeruginosa containing intracellular amidase (EC 3.5.1.4) activity. This enzyme catalyses the hydrolysis of aliphatic amides producing the corresponding organic acid (acrylic acid in the case of acrylamide) and ammonium ion (NH4+), which is detected by the selective electrode [13-15]. The biochemical reaction is shown in Fig. 1.
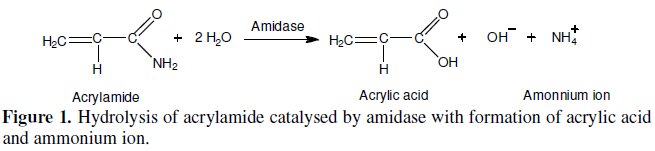
We strongly believe that this whole cell biosensor can be extremely useful in the detection and determination of acrylamide in food and environmental samples.
This monitorization is essential, regarding the reduction of risks for human health and environmental systems.
Experimental
Reagents and materials
Acrylamide solutions, of several concentrations, were prepared from commercial acrylamide, purchased from Merck.
Lactic acid, Himedia and ammonium sulphate, Merck were used in the preparation of bacterial growth media.
The solvent used in acrylamide solutions was TME (Tris-Mercaptoethanol- EDTA) 20 mM buffer pH = 7.2. This buffer was prepared with 20 mM Tris-HCl, Riedel-de Haën, 1 mM β-mercaptoethanol, Merck and 1 mM EDTA, Merck.
The solution used for the removal of unbound cells after each immobilization procedure was TMEGB (Tris-Mercaptoethanol-EDTA-Glycerol-Benzamidine) 50 mM buffer pH= 7.2. This buffer was prepared with 50 mM Tris-HCl, Riedelde Haën, 1 mM β-mercaptoethanol, Merck, 1 mM EDTA, Merck, 10 % (v/v) glycerol, Riedel-de Haën and 1 mM benzamidine, Merck.
This buffer was also used in the preparation of whole cells suspension (for immobilization purposes) and in the removal of ammonium ions after each catalytic reaction.
The various membranes used for whole cells immobilization were: modified polyethersulphone (Ultrabind, 0.45 μm), nylon (Whatman, 0.80 μm), polycarbonate (Nuclepore Track-Etch, 0.80 μm), cellulose acetate (Porafil, 0.80 μm), polyvinylidene fluoride (porablot, 0.20 μm), and nitrocellulose (Trans-Blot, 0.45 μm).
All solutions mentioned above were prepared daily with ultra pure (Milli-Q) water with a resistivity higher than 18.2 Wm, at 25 °C.
Bacterial strain and growth conditions
A constitutive mutant strain L10 of Pseudomonas aeruginosa was used as the source of amidase. Cells were grown overnight in a minimum salt media containing lactic acid (0.5%, v/v), and ammonium sulphate (0.1%, w/v) at pH 7.2 and at 37 °C, in an orbital shaker set to 250 rpm. The cultures were centrifuged at 7000 rpm for 10 min and the cells washed twice with saline. The resulting pellets were stored at -20 °C.
Instrumentation
The apparatus for the potentiometric assays consisted on an AUTOLAB PGSTAT12 potentiometer from Eco Chemie controlled with GPES v.4.9 software. The acquisition of the resulting data was accomplished by a personal desktop computer.
The working and reference electrodes used in all potentiometric measurements were respectively an ammonium ion selective electrode and the Ag/AgCl reference system.
Potentiometric measurements
All biosensing assays were carried out in a final volume of 5 mL of acrylamide 0.1 M solution, at room temperature, with magnetic stirring.
After each immobilization procedure of the biological recognition element, the biosensor was immersed in TMEGB 50 mM buffer for 15 min, with magnetic stirring, in order to remove any unbound cells of Pseudomonas aeruginosa.
The experimental setup for the potentiometric biosensor assays is shown in Fig. 2.
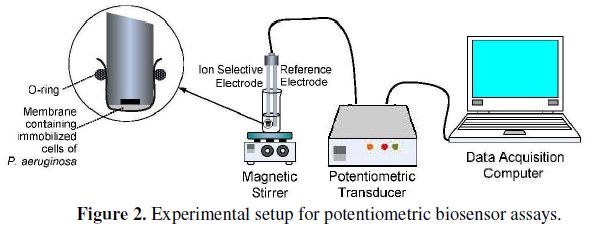
The biochemical reaction was recorded for a period of 6 min where the formation of NH4+, given by an increase in the potential values, could be followed. In fact, the endpoint potential reading of NH4+ formation was assumed to be a function of amidase activity. This parameter was considered as the biosensor's response and thus taken in consideration for all subsequent assays. All measurements were carried out in triplicates. A typical reaction profile can be observed in Fig. 3.
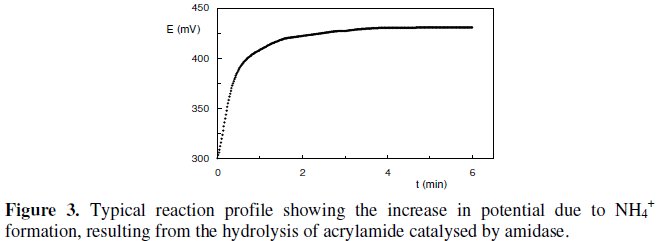
This reaction profile clearly shows the significant activity of the Pseudomonas aeruginosa intracellular amidase towards acrylamide by means of the constant slope established immediately after the beginning of the reaction. Also the potential difference measured only after 2 min of reaction time, states that there is not a diffusion resistance imposed by whole cells membrane. Therefore, substrate transport and assimilation into the cells are not rate-limiting processes for the whole cell biosensor.
Results and discussion
Immobilization of whole cells on single discs of polyethersulphone membrane The first approach considered for the immobilization of whole cells of Pseudomonas aeruginosa, was by using a solid support, namely single polyethersulphone (PES) membrane discs (∅ = 20 mm) [16-18]. In fact, a mixture of 60 μL cell suspension (in TMEGB 50 mM) with 5 μL of glutaraldehyde 5% (v/v), used as a cross-linking agent [19], was prepared in an eppendorf tube and immediately transferred to the center of the membrane disc. The mixture was then incubated at room temperature for 20 min, followed by membrane attachment to the surface of the ammonium electrode with an o-ring. Before the first assay, the biosensor was immersed in TMEGB 50 mM buffer for 15 min, with magnetic stirring, in order to remove any unbound cells of Pseudomonas aeruginosa.
Ammonium ion removal
Despite the fact that a good response was typically obtained with this biosensor (Fig. 3), a major drawback emerged from this procedure. In fact, the time needed for NH4+ complete removal from polyethersulphone membrane, after each catalytic reaction, so that the following assay could be initiated in identical conditions, was about 45 min. This assay was performed by monitoring the decrease in potential values, due to the removal of NH4+ from the membrane after each potentiometric assay (Fig. 4).
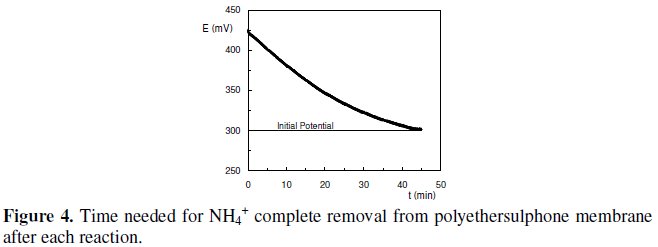
In this procedure, the biosensor was immersed in 5 mL of TMEGB 50 mM with magnetic stirring. The buffer was refreshed every 5 min.
In fact this period is not compatible with real time demanding measurements or on-line monitorizations, where most of the data collection is performed continuously [20-23].
Behaviour of other polymeric membranes
In order to overcome this problem, it was decided to extend this procedure to other polymeric membranes, such as cellulose acetate (CA) [24], nylon (NY) [25], polyvinylidene fluoride (PVDF) [26], nitrocellulose (NC) [27] and polycarbonate (PC) [28].
From these membranes, and according to several limitations observed, like low diffusion rate of the substrate, lack of signal or biosensor's response, extreme NH4+ removal periods, low ability for physical attachment to the electrode's surface due to unsuitable mechanical properties or high incubation periods, only nylon and polycarbonate membranes were considered for a comparative study with polyethersulphone. The results of the tests performed with these polymeric membranes are summarized in Table 1.
Comparative study between polyethersulphone, nylon and polycarbonate membranes
The main goal of this study was to find if nylon or polycarbonate membranes could be considered as an alternative to polyethersulphone (as immobilization support for whole cells of Pseudomonas aeruginosa), in what NH4+ removal time it concerns. One of the major concerns at this time was that the biosensor's signal or response obtained with polyethersulphone membranes wouldn't be compromised.
Whole cells immobilization procedure and further experimental conditions involving these assays are the same described in the sections concerning immobilization of whole cells on single discs of polyethersulphone membrane and potentiometric measurement, respectively.
Preliminary results showed that the biosensor's signal or response obtained with the new membrane materials were, in all aspects, similar to that verified with polyethersulphone membrane (Fig. 3).
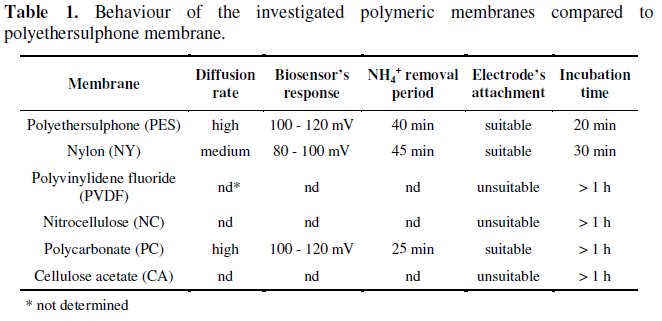
Regarding the time needed for complete removal of NH4+, the results presented in Fig. 5 show that polyethersulphone membranes needed about 40 min, whereas nylon membranes needed about 45 min, leading, from a global perspective, to a time consuming analysis.
On the other hand when using polycarbonate membranes the time needed for complete removal of NH4+ from the membrane decreased to about 25 min. Despite the fact that this value is about half of that found for the other two membranes, it is still significantly high, from a point of view where it is demanding to develop fast methods of analysis.
Double membrane biosensors
At this stage of the investigation, another limitation was observed, which resulted from the use of single membrane discs, where the biological element contacts directly with the electrode's surface. In fact, each time the biosensors were used and the membranes attached to the ammonium electrode, in order to perform the assays, and then removed for storage, it was observed that a significant amount of cells was lost. This observation was responsible for a premature decrease in the biosensor's overall activity, due to the loss of biological recognition element. In order to overcome this problem, instead of using single membrane disc, the hypothesis of using double membrane discs, in a âsandwichâ design, was considered. This way the cells would remain contained between the membranes, never contacting the electrode's surface, and thus prevent their premature loss. Consequently, the activity of the biosensors could be maintained for longer periods of time.
Prior to performing the assays involving the new membrane design, it was thought that the time needed for NH4+ removal from the double membranes could be twice the time observed for single discs. However, the obtained results, in Fig. 5, showed that no significant increase in this parameter was observed, when using double membranes.
In fact, the results obtained for single and double membranes were very similar, and once again polycarbonate double membrane exhibited the most favourable result in what NH4+ removal time it concerns.
At this time, since nylon membranes were the less suitable to this purpose, as well as in what biosensor's response it concerns (Table 1), only polyethersulphone and polycarbonate membranes were considered for further studies.
On the other hand, bearing in mind the limitations observed in the use of single membrane discs, as well as the coinciding results obtained for both designs, (Fig. 5), it was decided that only double membranes would be used in future assays.
Analytical performance of the biosensors for acrylamide detection The analytical characteristics regarding the performance of the biosensors using double membranes for acrylamide detection were evaluated and the results compared.
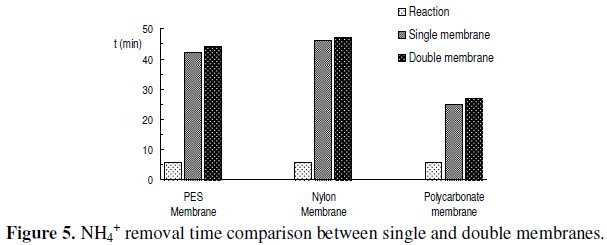
A calibration assay was carried out for each biosensor in the range of 1×10-5 M to 5×10-1 M of acrylamide concentration. The linear response found for the biosensor using polyethersulphone double membrane ranged from 1×10-3 M to 1×10-1 M, whereas for the biosensor using polycarbonate double membrane it was observed in the range of 5×10-4 M to 1×10-1 M. For both biosensors it was obtained a good correlation between the logarithm of acrylamide concentration and the variation in potential, E (mV). The Nernst equations obtained were the following:

A set of sensograms for both biosensors, representing the interval of linear response is shown in Fig. 6.
Once the calibration curves were obtained, the limit of detection (LOD) [30, 31] for acrylamide was determined for each biosensor. The biosensor using polyethersulphone double membrane exhibited a limit of detection of 6.31×10-4 M, whereas the value obtained for the biosensor using polycarbonate double membrane was 9.12×10-5 M.
The biosensors response times, considered as the time needed to achieve 90% of the endpoint response in a given reaction period [32], were 3.5 min for the polyethersulphone double membrane biosensor and 2.5 min for the polycarbonate double membrane sensor.
In order to compare the biosensors operating and storage stability with time, an assay involving the reutilization of the same membranes and consequently the same immobilized Pseudomonas aeruginosa cells, was carried out. These assays were performed once a week, according to the same experimental conditions described in the sections concerning immobilization of whole cells on single discs of polyethersulphone membrane and potentiometric measurements, respectively.
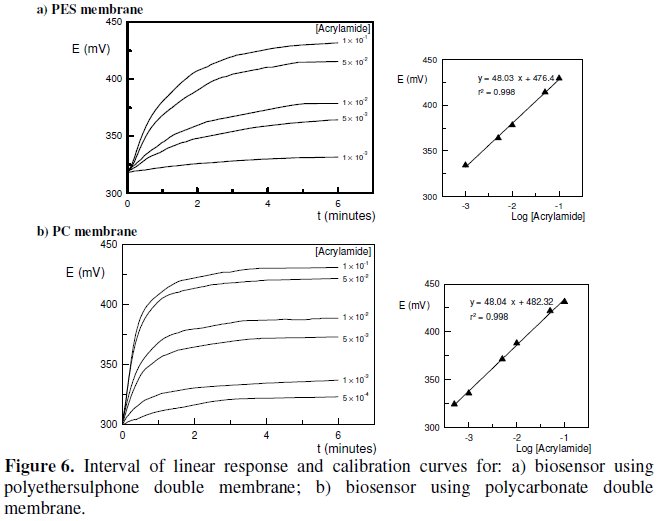
Between measurements, the membranes were stored at 4 °C in TMEGB buffer, pH = 7.2. The comparing results, in terms of the half-life time (t1/2), are shown in Fig. 7.
The half-life time of a quantity whose value decreases with time can be defined as the interval required for the quantity to decay to half of its initial value [16]. According to the results obtained in Fig. 7, the half-life time for the biosensor using polyethersulphone double membrane was about 27 days, whereas for the biosensor using polycarbonate double membrane it was only about 7 days. This poor result obtained with the later biosensor is due to some intrinsic limitations of the polycarbonate membrane that couldn't be overcome. In fact, a poor adherence of the cells to the membrane, and a low ability in assuming the transducer's shape, whenever it was attached to the electrode's surface, determined that a significant amount of cells was always prematurely lost, even when using a double membrane design. This observation was the main responsible for the reduced half-life time observed for the biosensor using polycarbonate membrane.
Table 2 resumes the figures of merit obtained for both biosensors, allowing an overall comparison between their behaviour and performance when using polyethersulphone and polycarbonate double membranes.
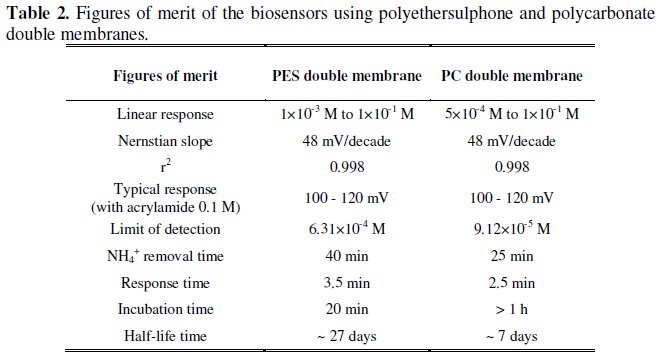
The results presented in Table 2 reveal some advantages of the biosensor with polycarbonate double membrane towards the biosensor with polyethersulphone double membrane. These advantages include a wider interval of linear response, a lower limit of detection and a shorter period for NH4+ removal. On the other hand, the incubation time observed for polycarbonate membrane is significantly higher than that for polyethersulphone. Also the half-life time obtained with polycarbonate membranes is only about 1/4 of the value observed for polyethersulphone.
At this stage it was taken in consideration that the main goals of the present investigation were the implementation of a simple and fast bioanalytical method for toxic amides determination, as well as the development of a device, with significant time stability, that could be reutilized as many times as possible. Bearing in mind these objectives, and considering the global results for both membranes, in particular the biosensors stability and the possibility of their reutilization, polyethersulphone was considered to be the most adequate membrane material for Pseudomonas aeruginosa cells immobilization, for the applications of the herein described electrochemical biosensor.
Conclusions and future developments
The present work involved the development and characterization of the merits and limitations of an electrochemical biosensor based on ion selective electrode and immobilized whole cells of Pseudomonas aeruginosa, with intracellular amidase activity, for the detection and determination of acrylamide.
The first approach to the biosensor's design involved the use of single discs of polymeric membranes as support for whole cells immobilization. However, this procedure determined a significant biomass loss, and a premature decrease in biosensor's overall activity.
Bearing in mind this limitation, a new approach was taken in consideration, and the behaviour of double membranes, in a ''sandwich'' design, was investigated. In fact, the use of double membranes was responsible for a significant reduction in biomass loss and thus enhancing biosensor's stability in time. Also, a major advantage that resulted from this procedure is that when double membranes were used, the immobilization of the cells, confined between both membranes, was performed without glutaraldehyde. This aspect is of significantly importance since the presence of glutaraldehyde increases the toxicity of the immobilization mixture containing the cells, compromising the biosensor's activity.
The biosensors using polyethersulphone and polycarbonate double membranes exhibited a good analytical performance, according to the figures of merit in Table 2. Typical responses of 100-120 mV, Nernstian slopes of 48 mV/decade and limits of detection in the order of 10-4 M and 10-5 M must be enhanced. However, the major problem encountered when using polymeric membranes in the immobilization process remained unsolved. In fact, the time needed for NH4+ removal after each biochemical reaction, couldn't ever be reduced below 25 min. On the other hand, this biosensor presents some advantages towards more classic methods for acrylamide determination, namely chromatography, since it involves lower costs, easy operation, simplified experimental design and apparatus, and enhanced specificity, since it is based on specific enzymatic reaction.
The referred characteristics allow us to believe that this biosensor may be significantly useful in the detection and determination of acrylamide in food and environmental analysis. The assessment of drinking and natural waters quality, in what acrylamide presence concerns, may be a preference field of application for this potentiometric biosensor, since accumulation and persistence in drinking water after treatment, groundwater and aquatic environments may occur.
Presently, in order to overcome the unique limitation, regarding the present work, which remains unsolved, we are investigating other immobilization matrices for whole cells of Pseudomonas aeruginosa as an alternative to the use of polymeric membranes. Bovine serum albumin (BSA), gelatine, nafion, agarose and sol-gel (both aqueous route using sodium silicate as a silica precursor and alkoxide route, using tetraethylorthosilicate), are examples of some materials that are being used and combined, to prepare different immobilization matrices.
In future work we will try to adequate the present biosensing system to another device, with reduced dimensions, with the same or a different transduction process (in particular condutimetric or piezoelectric). The performance of the biosensor in flow injection analysis will also be tested, regarding field and online applications. The development of an acrylamide biosensor using ''screen printing'' technology is one of the main future objectives. The possibility of using other microorganisms with intracellular amidase activity, as well as other amides, such as formamide or acetamide will also be investigated.
References
1. M. Friedman, J. Agr. Food Chem. 51 (2003) 4504. [ Links ]
2. Chia-Hsien Feng, Anal. Chim. Acta 684 (2010) 80. [ Links ]
3. J.M. Rice, Mutat. Res. 580 (2005) 3. [ Links ]
4. L. Brown, M. M. Rhead, D. Hill, K.C.C. Bancroft, Water Res. 16 (1982) 579. [ Links ]
5. M. Weiderborg, T. Källqvist, K.E. Ødergård et all, Water Res. 35 (2001) 2645. [ Links ]
6. C.-C. Wang, Æ C.-M. Lee, Curr. Microbiol. 55 (2007) 339. [ Links ]
7. S. Cavalli, S. Polesello, G. Saccani, J. Chrom. A 1039 (2004) 155. [ Links ]
8. F. Tateo, Int. J. Food Sci. Tech. 45 (2010) 629. [ Links ]
9. M. Anese, B. Quarta, J. Frias, Food Chem. 126 (2011) 435. [ Links ]
10. E. Susanna, Molec. Nutr. Food Res. 51 (2007) 239. [ Links ]
11. M. Pohankai, P. Skládal, J. Appl. Biomed.6 (2008) 57. [ Links ]
12. D. Grieshaber, R. MacKenzie, J. Vörös, E. Reimhult, Sensors 8 (2008) 1400. [ Links ]
13. S. Martins, A. Karmali, M. L. Serralheiro, Anal. Biochem. 355 (2006) 232. [ Links ]
14. J. Andrade, A. Karmali, M. A. Carrondo, C. Frazão, J. Biol. Chem. 282 (2007) 19598. [ Links ]
15. A.M.O. Brett, C.M.A. Brett, Electroquímica: Princípios, Métodos e Aplicações, Oxford University Press, 1996. [ Links ]
16. S.A.S.S. Gomes, M.J.F. Rebelo, Sensors 3 (2003) 166. [ Links ]
17. B.J. Ryan, C.O. Fágáin, BMC Biotech. 7 (2007) 1472. [ Links ]
18. A.J. Baeumner, N.A. Schlesinger, N. Slutzki et all, Anal. Chem. 74 (2002) 1442. [ Links ]
19. E. Akyilmaz, A. Yaoa, E. Dinçakaya, Anal. Biochem. 354 (2006) 78. [ Links ]
20. D.G. Bracewell, A. Gill, M. Hoare, Food Bioprod. Process 80(C2) (2002) 71. [ Links ]
21. G. Hanrahan, D.G. Patil, J. Wang, J. Environ. Monit. 6 (2004) 657. [ Links ]
22. H. Endo, Y. Yonemori, K. Hibi et all, Biosens. Bioelectron. 24 (2009) 1417. [ Links ]
23. B.C. Okeke, G. Ma, Q. Cheng et all, J. Microbiol. Meth. 68 (2007) 69. [ Links ]
24. V. Pundir, C.S. Pundir, J. Sci. Ind. Res. 67 (2008) 299. [ Links ]
25. M. Raje, R. Dhiman, S. Majumdar et all, Cytotechnology 51 (2006) 111. [ Links ]
26. T.-H. Young, J.-N. Lu, D.-J. Lin et all, Desalination 234 (2008) 134. [ Links ]
27. G. Singh, A. Bhalla, N. Capalash, P. Sharma, Indian J. Sci. Technol. 3 (2010) 48. [ Links ]
28. A.K. Basu, P. Chattopadhyay, U. Roychudhuri, R. Chakraborty, Biosens. Bioelectron. 21 (2006) 1968. [ Links ]
29. J. Janata, M. Josowicz, Solid State Ionics 94 (1997) 209. [ Links ]
30. E. Bakker, E. Pretsch, Trend. Anal. Chem. 24 (2006) 199. [ Links ]
31. D.L. Massart, B.G.M. Vandeginste, S.N. Deming et all, Chemometrics: A text book, Ed. B.G.M. Vandeginste, L. Kaufman, Elsevier, Amsterdam, 1988. [ Links ]
32. I. Uemasu, Y. Umezawa, Anal. Chem. 54 (1982) 1198. [ Links ]
Acknowledgements
The authors would like to acknowledge research grants (CIEQB 702 and PTDC/AGR-AAM/73460/2006) from Fundação para a Ciência e Tecnologia, Portugal. We are also grateful to Sonia Martins, Magda Semedo, Rita Pacheco, Dulce Gil and Sandra Silva for some technical help.
* Corresponding author. E-mail address: nsilva@deq.isel.ipl.pt
Received 11 February 2011; accepted 28 March 2011