Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Similars in
SciELO
Similars in
SciELO
Share
Revista de Ciências Agrárias
Print version ISSN 0871-018X
Rev. de Ciências Agrárias vol.41 no.3 Lisboa Sept. 2018
https://doi.org/10.19084/RCA17336
ARTIGO
Establishment of a genomic DNA extraction protocol for bamboo species
Estabelecimento de um protocolo de extração de DNA genómico para espécies de bambu
Rudieli M. Silva1* and Nathalia P. Ribeiro2
1 Departamento de Agricultura, Universidade Estadual Paulista – UNESP/FCA. Centro de Raízes e Amidos Tropicais – CERAT, Fazenda Experimental Lageado, Rua José Barbosa de Barros, 1780, CEP 18610-307, Botucatu, São Paulo, Brasil.
2 Departamento de Horticultura, Universidade Estadual Paulista – UNESP/FCA. Centro de Raízes e Amidos Tropicais – CERAT, Fazenda Experimental Lageado, Rua José Barbosa de Barros, 1780, CEP 18610-307, Botucatu, São Paulo, Brasil.
(*E-mail: rudielimds@gmail.com)
ABSTRACT
The bamboos Bambusa vulgaris and Guadua angustifolia are considered the most important species in South America. Generally, bamboo taxonomy is conducted by analysis of plants morphology, which sometimes may present morphological similarities, complicating the achievement of precise results. This fact can be changed by application of molecular biology methods; however, the Brazilian research of bamboo in molecular level is still limited. The Brazilian state of Mato Grosso do Sul has fragments of a natural bamboo forest with external characteristics similar to Guadua. In order to provide information about bamboo species, this paper evaluates the application of a modified protocol for DNA extraction from young leaves of two accesses of Guadua cultivated in Campo Grande/MS. It also evaluates the amplification efficiency of the matK and rbcL regions of the DNA isolated. It was possible to extract a mean of 487.5 ng μL-1 of DNA for both species with a purity level of 1.91. The molecular marker rbcL amplified the regions of 300 bp for both species, while the marker matK amplified the regions of 850 bp, also for both accesses. Results show that the modified protocol is useful for bamboos DNA extraction, with high concentration, quality, and able to be amplified by PCR.
Keywords: DNA extraction, Guadua angustifolia, molecular markers, polymerase chain reaction.
RESUMO
Os bambus Bambusa vulgaris e Guadua angustifolia são considerados as espécies mais importantes na América do Sul. Geralmente, a taxonômia de bambus é realizada pela análise da morfologia das plantas, que algumas vezes apresentam similaridades morfológicas, dificultando a obtenção de resultados precisos. Isso pode ser mudado pelo uso de métodos de biologia molecular, no entanto as pesquisas brasileiras com bamboo a nível molecular ainda é limitada. O estado do Mato Grosso do Sul possui fragmentos de florestas de bambu com características semelhantes ao Guadua. Com o intuito de fornecer informações sobre espécies de bambu, o presente trabalho avalia a aplicação de um protocolo modificado de extração de DNA de folhas jovens de duas amostras de Guadua cultivados em Campo Grande/MS. Também é avaliado a eficiência na amplificação das regiões matK e rbcL do DNA isolado. Foi possível extrair em média 487,5 ng μL-1 de DNA para ambas as espécies com nível de pureza de 1,91. O marcador molecular rbcL amplificou a região de 300 pb, enquanto que o marcador matK amplificou a região de 850 pb, ambos para os dois acessos. Resultados mostram que o protocolo modificado é útil para a extração de DNA de bambus, com alta quantidade, qualidade, e capaz de ser amplificado por PCR.
Palavras-chave: Extração de DNA, Guadua angustifolia, Marcadores moleculares, Reação da polimerase em cadeia.
INTRODUCTION
Bamboos are part of the Poaceae family, with approximately 1400 species already described within 651 genres distributed around the world (Behari, 2006). A great part of the bamboo species are original from Asian countries, however, they can be found both in tropical and temperate forests (Bystriakova et al., 2003).
Characteristics related to the facility of reproduction, high longevity, and productive level, make the bamboo wood commercially important for pulp paper industries, civil construction, and production of special furniture. In the Brazilian states of Maranhão and Paraíba are located the greatest productions of Bambusa vulgaris in the world, which is predominantly used for paper production (Neto et al., 2010). The authors also state that the charcoal obtained from species of B. vulgaris has more quality than the ones obtained from Eucalyptus.
Since the Brazilian colonial period, about 20 species of Asian bamboos were introduced into the Brazilian native forests (Silva et al., 2011). The Japanese immigration to Brazil was one of the processes that promoted the increase of Phyllostachys edulis in the country, species which has its bud largely consumed by people (Tombolato et al., 2012). Another important species is the Guadua sp. which presents a high-quality wood and can be found in the Amazonia and some parts of the Brazilian cerrado. Hence, Yeasmin (2015) claims that these species have adapted very well to the Brazilian climate characteristics, and have become economically important to the country.
An important fact related to bamboo usage in Brazil, is the Law number 12.484, of September 8th of 2011, which aims the increase of research with bamboo species in Brazil, as well as the stimulation of bamboo production by family farmers (Brazilian Government, 2011). According to the Brazilian Government (2011), bamboos are considered plants with a high agronomic potential, and then, are important for the internal economy.
According to the Brazilian Association of Bamboo Producers (2015), the Brazilian State of Mato Grosso do Sul presents more than 40 thousand hectares of native bamboo. According to the association, bamboo is a font of a sustainable economy, and it is economically viable for family farmers, once it is possible to produce bamboo for at least 70 years in the same area and being not necessary the use of heavy equipment for the harvest.
Brazil has about 1.5 million hectares of cultivated and native bamboo. The area of cultivated bamboo in Mato Grosso do Sul is still a small proportion of the total, mainly because of the lack of experiments with cultivated species. In Mato Grosso do Sul the native species is Guadua chacoensis, considered one of the best, commercially talking, among more than 1.3 thousand species (Brazilian Association of Bamboo Producers, 2015).
When bamboo species are considered as important commercial products, as it happens in Colombia and Brazil, it is necessary to establish methods of conservation for molecular characteristics of species (Kaneko et al., 2008). The knowledge of the genetic diversity of bamboo is essential for selection of good varieties for production (Blaxter et al., 2006), as it could happen in Mato Grosso do Sul.
According to Das et al. (2008), some molecular methodologies are already available for identification and characterization of bamboo species. However, it is difficult to establish the appropriate molecular method with the morphological taxonomy already used worldwide. The authors also state that molecular methods for bamboo taxonomy are already well developed, and highlight the use of RFLP (Restriction Fragment Length Polymorphism), RAPD (Randomly Amplified Polymorphic DNA), SCARs (Sequence Characterized Amplified Regions), AFLP (Amplified Fragment Length Polymorphism), SSRs (Simple Sequence Repeats or Microsatellites), and recently, the DNA barcoding. Although well developed, these molecular methods do not rely on DNA extraction protocols specific for bamboo species. Hence, it is important to establish a DNA extraction protocol that may be used as a standard by the researchers of this field.
The DNA barcoding is a technology already used in several animal and plant species to verify their molecular variability. This technology uses molecular markers to identify and amplify specific DNA regions (Little, 2011). In animal DNA barcoding, the researchers use the Cytochrome oxidase (COI), a protein-coding marker, as the DNA barcode standard, once it presents high copy number per cells and extensive length variation (Hollingsworth et al., 2011). In plants, a standard DNA barcode was not identified yet. Hence, some of the most studied markers in plants molecular biology are the maturase K (matK) and the ribulose-bisphosphate carboxylase (rbcL) (Biswal et al., 2012).
Although molecular methodologies for plants classification are already applied in the world, further investigation and the improvement of these methodologies used in bamboo taxa is still necessary. It is important to emphasize that the application of modern molecular techniques in bamboo species in order to identify and classify new species is necessary for our diversity conservation. However, the Brazilian research of bamboos in molecular level is still limited, mainly in relation to genomic DNA extraction protocols specific for bamboo species. Hence, this paper evaluates the efficiency of a protocol for DNA extraction of two bamboo accesses of Guadua cultivated in Mato Grosso do Sul and also evaluates the amplification of the regions rbcL and matK considered possible universal barcodes for plants.
MATERIAL AND METHODS
The methodology was conducted at the laboratory S-INOVA of the Universidade Católica Dom Bosco, Campo Grande, state of Mato Grosso do Sul.
Two accesses of cultivated bamboos were used for the experiments. Young leaves samples were collected at the São Vicente Research Institute (coordinates S20°2314 and W54°3629, at 532 m of altitude) which is responsible for the plant's maintenance. Both accesses were sold to the institute as being seedlings of Guadua, however, both of them presenting morphological differences (leaf and culm sizes). For this reason, the samples are named in this study as Guadua 1 and Guadua 2.
The protocol for DNA extraction
The extraction of DNA was conducted following the protocol established by Bonato et al. (2002) for DNA extraction of Stylosanthes spp. with modifications necessary for extraction of DNA from bamboo leaves.
Procedure:
Firstly, 5 g of leaf tissue was weighted, frozen in liquid nitrogen and ground to a fine powder. This powder was transferred to a 50 ml polypropylene tube with 10 ml of 1% CTAB buffer, incubated at 65 ºC for 60 minutes in a water bath, and then centrifuged at 4500 g for 10 minutes at 4 ºC. 225 μL of phenol and 225 μL of chloroform were added to the tube, which was gently shaken for 10 minutes and centrifuged at 4500 g for 10 minutes at 4 ºC. The supernatant was transferred to another tube and added, again, 225 μL of phenol e 225 μL of chloroform and repeated the step of centrifugation. The supernatant was transferred to another tube, and then, added 4.5 ml of a solution of chloroform and isoamyl alcohol at the proportion 24 to 1 (24 parts of chloroform and 1 part of isoamyl alcohol). The tube was gently shaken, centrifuged at 4500 g for 10 minutes at 4 ºC, and the supernatant transferred to another tube. The tube was treated with 600 μL of isopropyl alcohol (-20ºC) for DNA precipitation, and then, incubated at -20ºC for 30 minutes. The sample was centrifuged at 10000 g for 5 minutes at 4 ºC for the DNA pellet formation, and the supernatant carefully discarded. The DNA pellet was dissolved in 400 μL of TE buffer and precipitated by adding 20 μL of NaCl and 800 μL of ethanol absolute. The sample was centrifuged at 10000 g for 5 minutes at 4 ºC, and then the pellet washed with 70% ethanol (-20ºC). The ethanol was discarded and the DNA pellet dried at room temperature. Finally, the DNA pellet was dissolved in 50 or 100 μL of TE buffer, added 6 μL of RNAs end incubated at 37 ºC for 30 minutes and stored at -20ºC until use.
The DNA quantification was conducted using an Eppendorf Biophotometer. The genetic materials quality was observed by electrophoresis in 0.8% agarose gel, transferred into solutions of ethidium bromide for 15 minutes and distilled water for 10 minutes, and finally observed under UV light. The 1 kb plus DNA ladder was used as a pattern of molecular mass. All procedures, from DNA extraction to DNA quantification, were conducted in triplicate.
The CTAB buffer used for the extraction was prepared just before the beginning of the process. It was composed of 2% Cetyltrimethylammonium bromide, 100 mM Tris-HCl, 1% polyvinyl pyrrolidone, 20 mM EDTA and 1.4 M NaCl. The TE buffer was also prepared before the extraction process to be used fresh. This buffer is composed of 1 M Tris-HCl pH 8, 0.5 M EDTA pH 8, and distilled water.
The Polymerase Chain Reaction (PCR)
The PCR was conducted following the methodology described by New England Biolabs (2015). The amplification reactions were carried out using 100 ng μL-1 of genomic DNA added to 50 μL of PCR mix containing: 1.6 mM of MgCl, 0.2 mM of DNPTs, 5 μL of 1X Standard Taq Reaction Buffer, 1.5 U of Taq DNA Polymerase, 0.2 pmol primer. Two specific primers were used: rbcL (Forward: 5-ATGTCACCACAAACAGAGACTAAAGC-3; Reverse: 5-GTAAAATCAAGTCCACCRCG-3) and matK (Forward: 5CCTATCCATCTGGAAATCTTAG-3; Reverse: 5GTTCTAGCACAAGAAAGTCG-3).
The samples were transferred into a PCR thermocycler under the following conditions: initial denaturation at 94 ºC (5 min); 35 cycles of annealing at 94 ºC (1 min), 50 ºC (1 min) and 72 ºC (1 min); final extension at 72 ºC (10 min), followed by hold at 4 ºC infinite. The samples were stored into a fridge at -20 ºC overnight, and the amplified products were submitted to electrophoresis in 2% agarose gel, at same conditions as the DNA extracted was submitted for qualitys visualization.
RESULTS AND DISCUSSION
The mean concentration of DNA extracted from Guadua 1 and Guadua 2 were, respectively, 472 ng μL-1 and 503 ng μL-1. Results for DNA quantification are presented in Table 1. It is observed that the amount of DNA obtained from Guadua 2 is higher than that from Guadua 1, indicating that the concentration of DNA extracted by this protocol may vary according to the bamboo variety analyzed. Guadua 1 statistically differs from Guadua 2, however, both quantities of DNA, which must be at least in a range of 50 to 500 ng, are viable to the execution of the PCR reaction (Thermo Scientific, 2015). As observed, the DNA concentration for Guadua 2 is slightly higher than the specified, however, the Oxford Gene Technology (2016) explains that the amount and the concentration of DNA for use depends on the application and finality of the samples. The author also clarifies that all DNA samples can be diluted if necessary.
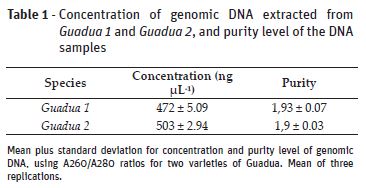
The genetic materials quality may also be observed in table 1 and figure 1. Table 1 represents the purity level of the DNA samples. Values of 1.93 and 1.90 were obtained for species Guadua 1 and Guadua 2 purity, respectively, in a ratio of A260/A280. It is important to say that these values must be in a range of 1.8 and 2.0. Values below than 1.8 mean that the samples are degraded by proteins, values over to 2.0 mean that the samples are degraded by RNA residues (Oxford Gene Technology, 2011). Figure 1 shows the electrophoretic profile of the DNA samples. The presence of a genomic DNA with good quality and concentration is notable.
The quality of the DNA obtained can also be related to the use of liquid nitrogen at the beginning of the extraction. According to Danner et al. (2011), the use of liquid nitrogen to macerate the young leaves can optimize and reduce the presence of undesired compounds in the final product. It happens mainly because the liquid nitrogen freezes the plant tissue, stopping the degradation and contamination of the DNA by phenolic compounds (Monsanto, 2016).
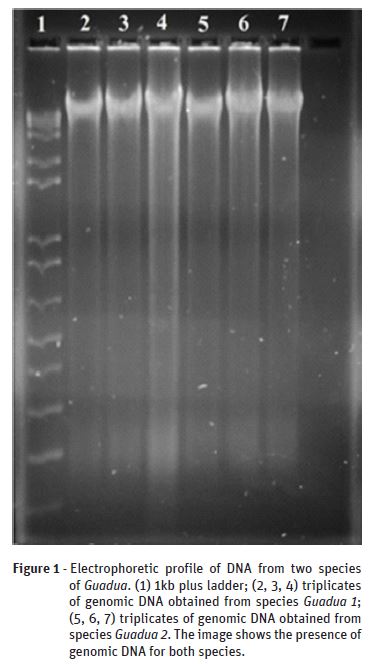
Figure 2 presents the electrophoretic profile of the amplified regions using rbcL and matK primers. It can be noticed that the rbcL primers amplified a region of 300 bp and the matK primers amplified a region of 850 bp, both primers for Guadua 1 and Guadua 2. The high similarity level presented by the electrophoresis analysis would suggest that there is low genetic variability between both Guadua species tested. However, the primers used do not distinguish different species only by their use, what means that further research needs to be carried out.
According to Li et al. (2011) matK is considered the most promising coding region in the genome DNA of plants. However, this primer presents difficulty in PCR amplification of non-angiosperms plants, a fact that complicates its use. In relation to the rbcL primer, the authors state that this gene region is of easy amplification, sequencing, and alignment, despite presenting low discriminatory power. That is why the combination of these two primers (matK + rbcL) was approved as universal barcodes for plants until the development of a single universal marker has been achieved. Hence, in the present work, it was found out that both primers can be used as molecular markers for bamboo species, once they proved efficient for bamboos DNA amplification. Also, the amplification efficiency of both regions confirmed that the DNA isolated by this modified protocol can be used for PCR analysis.
The increasing use of bamboo as an agricultural raw material in Brazil is evident in recent decades. The country is rich in diversity of bamboo species, and many of them present morphological similarities. Therefore, it would be important to the agricultural and biological sciences the development of efficient techniques of bamboo classification and differentiation through molecular biology, which is a limited research area in the country when it comes this plant species. Hence, this work becomes important, since it presents an efficient protocol for the extraction of DNA from bamboo species.
CONCLUSIONS
The modified protocol is viable for bamboo DNA extraction, presenting a high quality and concentration of genomic DNA for both bamboo species analyzed. Both of primers, rbcL and matK, are useful for DNA amplification of the two species analyzed, confirming that the DNA obtained by this methodology can be used for PCR analysis. Also, the primers rbcL and matK can be selected for further molecular analysis in bamboo species.
The use of liquid nitrogen is an efficient method to reduce the DNA degradation and the presence of undesired compounds in the final product.
REFERENCES
Behari, B. (2006) - Status of bamboo in India. Compilation of papers for preparation of national status report on forests and forestry in India. Survey and Utilization Division, Ministry of Environment and Forest, p. 109-120. [ Links ]
Biswal, D.K.; Debnath, M.; Kumar, S. & Tandon, P. (2012) - Phylogenetic reconstruction in the Order Nymphaeales: ITS2 secondary structure analysis and in silico testing of maturase K (matK) as a potential marker for DNA bar coding. BMC Bioinformatics, vol. 13, n. 17, p. 26. https://dx.doi.org/10.1186%2F1471-2105-13-S17-S26 [ Links ]
Blaxter, M.; Jenna, M.; Tom, C.; Fran, T.; Claire, W.; Robin, F. & Eyualen, A. (2006) - Defining operational taxonomic units using DNA barcode data. Philosophical Transactions of the Royal Society B, vol. 360, n. 1462, p. 1935-1943. https://dx.doi.org/10.1098%2Frstb.2005.1725 [ Links ]
Bonato, A.L.V.; Verzignassi, J.R.; Resende, R.M.S.; Fernandes, C.D. & Leguizamon, G.O.C. (2002) - Extração de DNA genômico de Stylosanthes spp. Campo Grande: Embrapa Gado de Corte, Comunicado Técnico 78, 2002. 4p. [cit. 2015-05-08]. <http://old.cnpgc.embrapa.br/publicacoes/cot/pdf/COT78.pdf> [ Links ].
Brazilian Association of Bamboo Producers (2015) - O bambu é uma alternativa paras recuperação as áreas degradadas nas propriedades rurais de MS. Campo Grande: Senar MS. [cit. 2015-06-15]. <http://senarms.org.br/com-40-mil-hectares-ms-possui-a-3a-maior-area-de-bambu-nativo-do-brasil/> [ Links ].
Brazilian Government (2011) - Law number 12.484, of September 8th 2011. Brasília: Presidência da República. [cit. 2015-06-10]. <http://www.planalto.gov.br/ccivil_03/_ato2011-2014/2011/lei/l12484.htm>.
Bystriakova, N.; Kapos, V.; Lysenko, I. & Stapleton, C. (2003) - Distribution and conservation status of forest bamboo biodiversity in the Asia‐Pacific region. Biodiversity and Conservation, vol. 12, n. 9, p. 1833–1841. https://doi.org/10.1023/A:1024139813651 [ Links ]
Danner, M.A.; Sasso, S.A.Z.; Bittencourt, J.V.M.; Citadin, I. & Sachet, M.R. (2011) - Protocol suggested for DNA extraction of jabuticaba trees. Ciência Florestal, vol. 21, n. 2, p. 363-367. http://dx.doi.org/10.5902/198050983241 [ Links ]
Das, M.; Bhattacharya, S.; Singh, P.; Filgueiras, T.S. & Pal, A. (2008) -Bamboo taxonomy and diversity in the era of molecular markers. Advances in Botanical Research, vol. 47, p. 225-268. https://doi.org/10.1016/S0065-2296(08)00005-0 [ Links ]
Hollingsworth, P.M.; Graham, S.W. & Little, D.P. (2011) - Choosing and using a plant DNA barcode. PLoS One, vol. 6, n. 5, art. e19254. https://doi.org/10.1371/journal.pone.0019254 [ Links ]
Kaneko, S.; Franklin, D.C.; Yamasaki, N. & Isagi, Y. (2008) - Development of microsatellite markers for Bambusa arnhemica (Poaceae: Bambuseae), a bamboo endemic to northern Australia. Conservation Genetics, vol. 9, n. 5, p. 1311–1313. https://doi.org/10.1007/s10592-007-9467-z [ Links ]
Li, F.W.; Kuo, L.Y.; Rothfels, C.J.; Ebihara, A.; Chiou, W.L.; Windham, M.D. & Pryer, K.M. (2011) - rbcL and matK earn two thumbs up as the core DNA barcode for ferns. PLoS One, vol. 6, n. 10, p. e26597. https://doi.org/10.1371/journal.pone.0026597 [ Links ]
Little, D.P. (2011) - DNA barcode sequence identification incorporating taxonomic hierarchy and within taxon variability. PLoS One, vol. 6, n. 8, art. e20552. https://dx.doi.org/10.1371%2Fjournal.pone.0020552 [ Links ]
Monsanto (2016) - A recommended procedure of DNA extraction from plant tissues. Monsanto. [cit. 2016-05-08]. <http://www.monsanto.com/products/documents/dna-detection/dna_im.pdf> [ Links ].
Neto, M.C.L.; Neto, E.B.; Barreto, L.P. & Silva, J.A.A. (2010) - Exportation of macronutrients in commercial growth of bamboos in the state of Paraíba. Revista Árvore, vol. 34, n. 2, p. 251-257. [ Links ]
New England Biolabs (2015) - PCR protocol for taq polymerase with standard taq buffer. New England Biolabs. [cit. 2015-05-10]. <https://www.neb.com/protocols/1/01/01/taqdnapolymerasewithstandardtaq-bufferm0273> [ Links ].
Oxford Gene Technology (2011) - Understanding and measuring variations in DNA sample quality. Oxford Gene Technology. [cit. 2016-05-08]. <http://www.ogt.com/resources/literature/483_understanding_and_measuring_variations_in_dna_sample_quality> [ Links ].
Silva, I.F.; Pereira, D.S. & Silva, S.R.F. (2011) - Morphological studies of bamboo (Bambusa cf. vulgaris L.), an invasive species in the Atlantic forest on the Maceió Municipal Park- Alagoas. Revista Semente, vol. 6, p. 99-109. [ Links ]
Thermo Scientific (2015) - Assessment of nucleic acid purity. Thermo scientific. [cit. 2015-05-23]. <http://www.nanodrop.com/Library/T042-NanoDrop-Spectrophotometers-Nucleic-Acid-Purity-Ratios.pdf> [ Links ].
Tombolato, A.F.C.; Greco, T.M. & Pinto, M.M. (2012) - Ten exotic species of bamboo more common in Brazilian landscape projects. Revista Brasileira de Horticultura Ornamental, vol. 18, p. 105-114. [ Links ]
Yeasmin, L.; Ali, M.D.; Gantait, S. & Chakraborty, S. (2015) - Bamboo: an overview on its genetic diversity and characterization. 3 Biotech, vol. 5, n. 1, p. 1-11. https://dx.doi.org/10.1007%2Fs13205-014-0201-5 [ Links ]
ACKNOWLEDGEMENTS
To the S-INOVA laboratories of molecular biology of the Universidade Católica Dom Bosco where the experiments were conducted, especially to the Professor Carina Elisei de Oliveira for the help in the analyses. The authors also thank the CeTeAgro, Centro de Tecnologia e Análises do Agronegócio, in special the Professor Marney Pascoli Cereda for granting us all Guadua leaf samples for the DNA extraction procedure.
Received/recebido: 2017.12.21
Received in revised form/recebido em versão revista: 2018.02.27
Accepted/aceite: 2018.03.01