Serviços Personalizados
Journal
Artigo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Acessos
Acessos
Links relacionados
-
 Similares em
SciELO
Similares em
SciELO
Compartilhar
Ciência e Técnica Vitivinícola
versão impressa ISSN 0254-0223
Ciência Téc. Vitiv. v.25 n.2 Dois Portos 2010
Restriction profiles of 26S rDNA as a molecular approach for wine yeasts identification
Geni C. Zanol *, M. Margarida Baleiras-Couto, Filomena L. Duarte
Instituto Nacional de Recursos Biológicos, I.P./ INIA Dois Portos, Quinta dAlmoínha 2565-191, Dois Portos, Portugal
SUMMARY
The complex microbial ecosystem existing in grape, must and wine comprises a wide diversity of yeast species. The knowledge of composition and dynamics of yeast biota occurring along vinification process would provide a better control of wine quality. The sequence of D1/D2 domain of 26S ribosomal DNA (rDNA), reflects ascomycetous yeast phylogenetic relationships and enables their separation at the species level. A region of the 26S rDNA, with around 1100 bp comprising domain D1/D2, was amplified by PCR and then digested with restriction endonucleases (ApaI, HinfI, MseI, HaeIIIand CfoI) in order to differentiate yeast species frequently isolated from grape surfaces, wine and cellar equipments. A total of 78 yeast strains (including 36 type strains) belonging to 53 species were used to generate the restriction profiles. Numerical analysis of the profiles generated by the five restriction enzymes enabled to group the strains in 47 different clusters and 42 of them clearly corresponded to different yeast species. The remaining groups comprise closely related species. The enzymes MseI, HaeIIIand CfoIrevealed a high discrimination power and the restriction profiles generated were sufficient to clearly identify the 42 species mentioned above. Despite one of the clusters included different yeast genera, with different wine characteristics, the common wine spoilage yeasts Zygosaccharomyces bailii and Z. lentus could be separatedto one distinctive cluster through the use of ApaI restriction profiles. Since the analysis of restriction profiles of amplified 26S rDNA showed to be a valuable method to identify oenological yeast species, a database comprising the majority of wine yeast biota was created to be applied both at research and industrial environment.
Key words: 26S rDNA, endonucleases, non-Saccharomyces yeasts, restriction profiles, wine yeasts
Identificação de leveduras de interesse enológico por perfis de restrição do ADNr 26S
RESUMO
O ecossistema microbiano existente nas uvas, no mosto e no vinho é composto por uma grande diversidade de espécies de leveduras. O conhecimento deste biota de leveduras ao longo do processo de vinificação permite um melhor controlo da qualidade do vinho. Para a identificação de leveduras o ADN ribossómico (ADNr) tem-se revelado muito adequado para estimar relações filogenéticas, consideradas pelas correntes mais actuais da taxonomia como estando na base da classificação taxonómica. No presente trabalho, avaliou-se um método baseado na amplificação do ADNr 26S, compreendendo a região D1/D2, seguido de digestão por enzimas de restrição - Perfis de Restrição - para a identificação de espécies de leveduras envolvidas no processo de produção de vinho. Esta avaliação foi efectuada através do uso de 78 estirpes pertencentes a 53 espécies (incluindo 36 estirpes tipo). Utilizaram-se as enzimas de restrição ApaI, HinfI, MseI, HaeIIIe CfoI e, análise numérica dos perfis de restrição gerados permitiu agrupar as espécies estudadas em 47 grupos, 42 dos quais correspondendo a uma única espécie. As enzimas de restrição MseI, HaeIII e CfoI foram as que apresentaram maior poder discriminante ao nível da espécie, permitindo a identificação das mesmas 42 espécies. Apesar da enzyma ApaIter apresentado o mais baixo grau de polimorfismo, esta enzima poderá ser útil para medidas de controlo uma vez que seu perfil de restrição pôde agrupar em um grupo distinto as leveduras Zygosaccharomyces bailii e Z. Lentus. O método desenvolvido revelou eficácia, rapidez e facilidade de aplicação na identificação de leveduras de interesse enológico. Com o presente trabalho iniciou-se a construção de uma base de dados de perfis de restrição para posterior aplicação em condições industriais e de investigação.
Palavras-Chave: ADNr 26S, endonucleases, leveduras enológicas, leveduras não-Saccharomyces, perfis de restrição
INTRODUCTION
The art of wine making represents one of the oldest technological uses of yeast by man. Only during the last century the scientific knowledge of wine has significantly increased, assisted by newly developed techniques that permitted deeper investigation into the biological and physiological diversity of yeast species associated to the process (Pretorius, 2000). The grape has an incontestable influence in aroma and flavour of wines leading to the creation of distinct products. However, wine has more flavour than the grape juice which is fermented from (Romano et al., 2003) and it is the metabolism of grape constituents by yeast that is essential to the development of wine flavour (Bartowsky and Pretorius, 2009).
The wine fermentation is a complex ecological and biochemical process involving the sequential development of different microbiota such as non-Saccharomyces yeasts, Saccharomyces yeasts and lactic acid bacteria present in must and on surface of cellar equipments (Fleet, 2003). The non-Saccharomyces yeasts can produce a diversity of enzymatic activities and fermentation metabolites of oenological importance and may interfere with the growth and/or change the fermentation behaviour of the starter Saccharomyces cerevisiae yeast, thus ultimately influence wine quality (Cabrera et al. 1988; Romano et al., 1997; Ciani and Ferraro, 1998; Ciani and Maccarelli, 1998; Ferreira et al., 2001; Romano et al., 2003; Ciani et al., 2006; Domizio et al., 2007; Bely et al., 2008; Romano et al., 2008). The use of selected strains of S. cerevisiae as starters became a widespread practice in wineries. Nevertheless, wine makers have recently returned to spontaneous fermentation as well as to the use of non-Saccharomyces in order to obtain wine of distinctive quality and diversified products.
On the other hand, yeasts can negatively affect wine quality. Spoilage yeasts such as Brettanomyces/Dekkera produce volatile phenols and acetic acid that under uncontrolled conditions can lead to sensorial defects (Renouf and Lonvaud-Funel, 2007). Zygosaccharomyces is another yeast genus that is often regarded as synonymous of food spoilage due to their osmotolerance and resistance to food preservatives (Loureiro and Malfeito-Ferreira, 2003). Therefore, the analysis and identification of yeast biota throughout wine fermentation and conservation are currently important driving forces for innovation in wine technology.
Traditionally, yeast taxonomy has been based on morphological, physiological and biochemical characteristics of species and genera which ambiguity due to strain variability has led to errors in classification (Martini, 1992; Kurtzman and Robnett, 1994; Kurtzman and Fell, 1998). Isoenzymes electrophoretic profiles have also been applied and prove to reflect DNA based yeast species delimitation (Smith et al., 1990; Duarte et al., 1999; Sampaio et al., 2001; Naumova et al., 2003; Duarte et al., 2004). However, this technique is highly time-consuming.
Several approaches based on nucleic acids polymorphisms have been developed in an attempt to simplify yeast identification, such as electrophoretic karyotyping, temperature gradient gel electrophoresis (TGGE), microsatellite PCR fingerprinting, random amplified polymorphic DNA, ribosomal DNA (rDNA) restriction profiles and partial rDNA sequencing (Török et al.,1993; Baleiras-Couto et al., 1995; Baleiras-Couto et al., 1996; Guillamón et al.,1998; Kurtzman and Robnett, 1998; Esteve-Zarzoso et al., 1999; Hernán-Gómez et al., 2000; Esteve-Zarzoso et al., 2003; Baleiras-Couto et al., 2005; Rodriguez et al., 2010).
Nowadays, innovative wine yeast identification techniques such as DGGE (Denaturing Gradient Gel Electrophoresis) on PCR amplified rRNA genes, FISH (Fluorescence in situ Hybridization), real time quantitative PCR (qPCR) and next-generation DNA sequencing can enable the quantification and/or to monitor yeast dynamics throughout the fermentation process (Hierro et al., 2007; Mardis, 2008; Salinas et al., 2009; Tessonniere et al., 2009; Zott et al., 2010). However, these techniques need sophisticated and expensive equipments which are not commonly available.
The ribosomal genes (5.8S, 18S and 26S), which have as ultimate function the protein synthesis, are grouped in tandem forming transcription units that are repeated in the genome (Fernández-Espinar et al., 2006). rRNA genes have a common origin, are present in all cellular organisms and have proved to be adequate to establish taxonomic relationships, namely on yeasts, as it is present in all cellular organisms, have a common origin and are easy to sequence (Kurtzman and Pikur, 2005). Nucleotide sequences of the D1/D2 domains of the large subunit (26S) of rDNA are sufficiently substituted to allow recognition of most individual yeast species. Kurtzman and Robnett (1998) have sequenced D1/D2 domains for all known ascomycetous yeasts thus, initiating a universal database for rapid identification.
Simpler identification methods were developed based on the amplification of specific regions of rDNA followed by restriction of the amplified fragment. The digested fragments are then separated by electrophoresis in agarose gels and their sizes determined by comparison with appropriate markers. White et al. (1990) used this methodology to amplify the ribosomal gene 5.8S and the adjacent intergenic regions ITS1, ITS2 and further to digest with restriction enzymes. Another ribosomal region that is very useful to differentiate at species level is the one that includes 18S gene and the intergenic region ITS1 (Baleiras-Couto et al., 1996; Dlauchy et al., 1999). Since then, this approach has been used for identifying yeast species mainly associated alcoholic beverages and soft drinks (Guillamón et al., 1998; Esteve-Zarzoso et al., 1999; Arias et al., 2002; Ferreira et al., 2009). Restriction profiles generated have been considered reproducible, cheaper, a less-laborious method and frequently used for yeast identification (Fernández-Espinar et al., 2006).
Baleiras-Couto et al. (2005) started to evaluate the restriction profiles of a PCR amplicon of the large subunit of rDNA (26S rDNA), comprising the D1/D2 region, as a routine methodology to examine wine yeast species. In the present study, we extended the restriction profiles, originated through digestion with five restriction enzymes (ApaI, HinfI, MseI, HaeIIIand CfoI), of the same PCR amplicon, in order to develop an efficient and rapid methodology for oenological yeasts genotyping. The aim of this work was to create a database of restriction profiles, based on certified yeast strains, to be used in wine related yeast identification carried out both at research and industrial level.
MATERIAL AND METHODS
Microorganisms
A total of 78 yeast isolates, comprising 53 species belonging to 22 genera, included in the Colecção de Microrganismos EVN (INRB/INIA Dois Portos), were used in the present study (Table I). Thirty eight strains were originated from other culture collections, 36 of which are type strains. The remaining 40 strains were isolated from grapes, wine and cellar equipments in our Laboratory and identified by DNA sequencing of D1/D2 region of rDNA.
Strains used in the present study, their collection number, geographical origin and sources of isolation (where available).
Estirpes de leveduras utilizadas no presente trabalho e respectivos números de colecção, origem geográfica e fonte de isolamento (quando disponíveis).
Yeast cells were grown on YPD medium (20g/L D-glucose, 10 g/L bacto-peptone, 5 g/L yeast extract and 20 g/l agar) for 48 to 72 hours at 25ºC. Two to three loops of yeast culture (from fresh YPD agar plates) were resuspended in 500 µl of ultrapure sterilised water. Yeast cells lysate was obtained by disrupting cells through freezing of cell suspension in liquid nitrogen for 5 min, followed by incubation at 95 ºC for 5 min, accordingly to Baleiras-Couto et al. (2005). The cell lysate containing DNA was then used for PCR amplification purposes. When the ribosomal DNA amplification by PCR was not successful, the cells lysate was obtained by cell disruption using glass beads (0.5 mm Ø) in 500 µl lyses buffer (50 mM Tris-HCl, 250 mM NaCl, 50 mM EDTA and 0.3 % SDS). The cell lysate solution was appropriately diluted and then used for PCR amplification.
Amplification of the ribosomal DNA D1/D2 region
Primer sequences for the amplification of 26S rDNA fragments were as follows: NL1 (5-GCATATCAATAAGCGGAGGAAAAG-
Restriction analysis
Aliquots (3-10 μL according to the band intensity) of PCR products were digested with 3 U and 5 U, respectively, of restriction enzymes MseI, HinfI and ApaI (MBI Fermentas) and HaeIII and CfoI (Promega, Madison, WI) in a final volume of 20 μL, following manufactures instructions. The resulting fragments were separated by 2% agarose gel electrophoresis followed by ethidium bromide staining, as referred above. A standard DNA marker (100 bp DNA Ladder, MBI Fermentas) was used as a reference to determine the size of digested fragments. Restriction fragments were visualised under UV light and digital images were acquired through a Kodak 290C camera and processed as referred above. . All restriction profiles obtained were analysed using GelCompar II software, version 5.1 (Applied Maths,Saint-Martens-Latem, Belgium) which determined the molecular sizes of restriction products. Fragments smaller than 100 bp were not included on the analysis because of their low reproducibility. Similarities among banding profiles of the strains in study were based on Dice coefficient and dendrograms were generated by the Unweighted Pair Group Method using Arithmetic Average (UPGMA) clustering algorithm.
RESULTS AND DISCUSSION
Several molecular methods are presently being applied for microbiological identification and classification. Each method has its advantages and disadvantages according to the convenience of applicability, reproducibility, availability of equipments, and resolution level.
In this study, analysis of restriction profiles of NL1-LR6 region of 26S rDNA was used to differentiate wine yeast species associated to wine production. In a total of 78 strains comprising 53 species, the PCR amplification yielded a fragment size of around 1100-1150 bp. The amplified fragment was then digested with five endonucleases (ApaI, HinfI, MseI, HaeIIIand CfoI) and the restriction products were separated by agarose gel electrophoresis. Representative restriction profiles presented by the 53 yeast species analysed, are shown in Figure 1.
Figure 1 – 53 yeast species representative restriction profiles obtained after digestion with ApaI, HinfI, MseI, HaeIII and CfoI enzymes of the 26S rDNA region. The number following each species corresponds to the access number of Colecção de Microrganismos EVN (INRB/INIA Dois Portos); (T), (NT) and (LT) mean type, neotype and lectotype yeasts, respectively.
Perfis de restrição representativos das espécies de 53 espécies de leveduras obtidos após digestão de uma região do ADNr 26S com as enzimas ApaI, HinfI, MseI, HaeIII and CfoI. O número que segue a espécie de cada estirpe corresponde ao número de entrada na Colecção de Microrganismos EVN (INRB/ INIA Dois Portos); (T), (NT) e (LT) significam leveduras tipo, neotipo e lectótipo, respectivamente.
Each restriction enzyme generated a large number of digested fragments (19 or 20), with exception of ApaI which originated only 10 band classes, allowing the discrimination of only four species (Table II). Indeed, for most analysed yeast species (36), this enzyme was not able to digest the PCR amplified fragment, a fact that was already reported by Baleiras-Couto et al. (2005). The profiles generated after digestion with ApaI enzyme presented the lowest polymorphism and discrimination power.
TABLE II.
Characteristics of the restriction fragment length polymorphism profiles of the PCR amplified 26S rDNA region corresponding to each restriction enzymes ApaI, HinfI, MseI, HaeIIIand CfoI.
Características dos perfis de restrição gerados após digestão com cada uma das enzimas ApaI, HinfI, MseI, HaeIII and CfoI do produto amplificado por PCR da região 26S do ADNr.
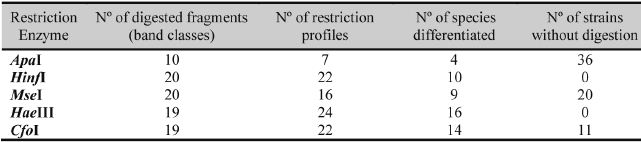
On the other hand, the digestion with restriction enzymes HaeIIIand CfoI produced higher number of well-developed bands and higher degree of polymorphism (with 22 and 24 distinct restriction profiles, respectively). The discrimination power of HaeIIIand CfoI was also higher as many restriction profiles were species specific (16 and 14 respectively). The remaining enzymes HinfI and MseI despite the high degree of polymorphism (with 22 and 16 restriction profiles, respectively) showed an intermediate discrimination power presenting high number of profiles shared by many of the studied species.
Cluster analysis of the strains in study were performed considering the fingerprints of all restriction enzymes, their relationship was calculated by applying the Dice coefficient, and a dendrogram was generated using UPGMA clustering algorithm. The 26S rDNA-based restriction analysis generated 47 clusters 42 of them corresponding to a single yeast species and only five clusters not species-specific (Figure 2). The calculated cophenetic correlation coefficient (0.83) indicates a good fit for the cluster analysis. The species-specific restriction profiles generated by the five endonucleases used in this study allowed the identification of the most predominant non-Saccharomyces yeast genus found in grape surfaces or winery environments such as Hanseniaspora, Candida, Pichia, Rhodotorula, and Kluyveromyces (Longo et al., 1991; Fleet and Heard, 1993; Schütz and Gafner, 1993; Torija et al., 2001; Clemente-Jimenez et al., 2004; Zott et al., 2008). This identification is of major importance as non-Sacharomyces yeasts might influence wine fermentations both directly, through production of off-flavors, and indirectly by modulating the growth or metabolism of the dominant Saccharomyces population (Fleet, 2003).
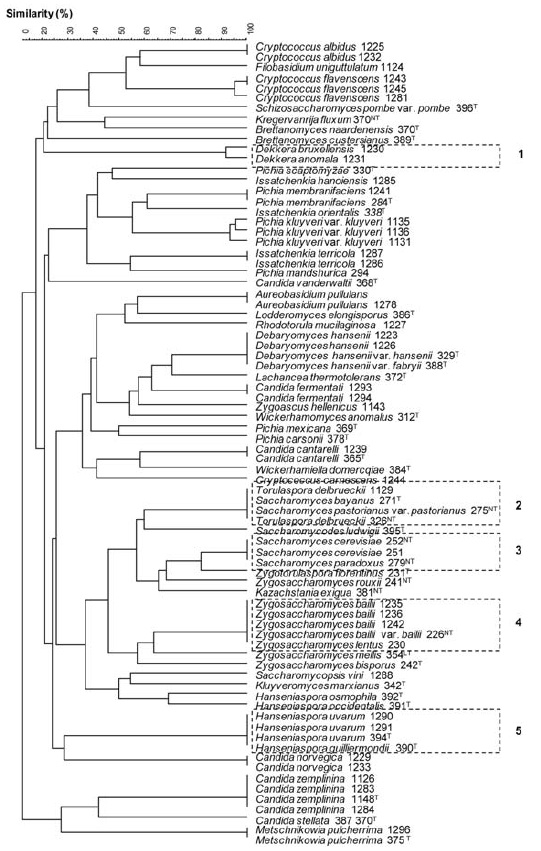
Figure 2 - Dendrogram of restriction profiles fingerprint, obtained after digestion with HinfI, MseI, ApaI, HaeIII and CfoI enzymes, presented by the 78 yeast strains. Dendrogram was generated by the Unweighted Pair Group Method using Arithmetic Average (UPGMA) clustering algorithm, calculated by using GelCompar II (version 5.1), cophenetic correlation coefficient = 0.83. The five clusters that could not be solved at species level are shown in dotted lines. The number following each species corresponds to the access number of Colecção de Microrganismos EVN (INRB/INIA Dois Portos); (T), (NT) and (LT) mean type, neotype and lectotype yeasts, respectively.
Dendrograma representando a semelhança entre as 78 estirpes com base nos perfis de restrição de ApaI, HinfI, MseI, HaeIII and CfoI obtidos de uma região do ADNr 26S. O dendrograma foi criado usando o coeficiente de Dice pelo método de agrupamento UPGMA (GelCompar II, versão 5.1), coeficiente de correlação cofenética = 0,83. Os cinco grupos em que não foi possível a identificação a nível de espécies estão indicados por linhas pontilhadas. O número que segue a espécie de cada estirpe corresponde ao número de entrada na Colecção de Microrganismos EVN (INRB/INIA Dois Portos); (T), (NT) e (LT) significam leveduras tipo, neotipo e lectótipo, respectivamente.
The non-Saccharomyces yeast species belonging to Metschnikowia, Kluyveromyces, Cryptococcus, Rhodotorula, Aureobasidium, Issatchenkia, Debaryomyces, Lachancea, Zygoascus and Saccharomycodes genera were all well assigned by presenting distinctive restriction profiles (Figure 2). These yeast species although in a lower extent, are normally present during wine fermentation (Mills et al., 2002; Baleiras-Couto et al., 2005; Nisiotou et al., 2007; Bisson and Joseph, 2009).
Schizosaccharomyces pombe, characterized by its special mode of vegetative reproduction and a certain degree of osmophily, can cause food spoilage (Esteve-Zarzoso et al., 1999). This species presented a unique restriction profile and, therefore, could be clearly separated.
In the present study, unique species-specific restriction profiles for the five studied Pichia species were obtained (Figure 2). Some Pichia species are present at high levels at the beginning of fermentations and have been associated with the development of surface flora in wines exposed to air or incompletely filled tanks or barrels (Fleet, 1993). The P. membranifaciens species may also present killer property by producing toxins that could inhibit the growth of some spoilage yeast such as Brettanomyces bruxellensis (Santos et al., 2009).
The very heterogeneous genus Candida includes all yeast species that cannot be classified in any other assexual ascomycetous yeast genera (Esteve-Zarzoso et al., 1999). Some Candida species have become very interesting for oenology due to their highly fructophilic nature allowing their use along with S. cerevisiae which is highly glucophilic (Mills et al., 2002). In this study, we analysed six Candida species that are frequently isolated in food and beverages. Through the restriction profiles generated with the five endonucleases, all these species could be clearly assigned (Figure 2). In some cases, Candida species have been shown to be able to complete the alcoholic fermentations (Clemente-Jimenez et al., 2004). The species C. stellata was found to be present at high level in musts (Hierro et al., 2006; González et al., 2007). However, in a recent work Csoma and Sipiczki (2008) have proposed that most isolates from grapes and wine are C. zemplinina rather than C. stellata. In this work, both species were evidently separated (Figure 2).
The unsolved group, constituted by Dekkera anomala and D. bruxellensis, which presented identical restriction profiles, was clearly separated from all other studied species constituting a reliable approach for Dekkera genus identification (cluster number 1, Figure 2). While Esteve-Zarzoso and co-authors (1999) clearly separated these two species using the 5.8S-ITS region restriction profiles, these authors could not separate D. anomala from H. uvarum and H. guilliermondii. In an industrial perspective, the methodology under study enabled the identification of the genus Dekkera which includes dangerous wine spoilers as they negatively modify physical and sensorial properties of wine provoking severe economical losses (Loureiro and Malfeito-Ferreira, 2003). The closer Bretanomyces species (B. naardenensis and B. custersianus) were also separated from each other and from Dekkera species.
The species Torulaspora delbrueckii, Saccharomyces bayanus and S. pastorianus were grouped in one cluster whereas S. cerevisiae and S. paradoxus were separated from them forming another cluster (clusters number 2 and 3, Figure 2). T. delbrueckii can produce positive effects on the taste and aroma of wines (Ciani and Maccarelli, 1998) whilst Saccharomyces complex (S. bayanus, S. cerevisiae, S. paradoxus and S. pastorianus) is the most strongly fermenting and ethanol-tolerant yeast group which takes over the wine fermentation (Fleet and Heard, 1993). In an early study, James and co-authors (1997) reported that the four species of the Saccharomyces sensu stricto were found to be closely related, displaying sequences similarity of the 18S rDNA higher than 99.9 %. Indeed, formerly the separation of Saccharomyces sensu stricto species could be achieved through isoenzyme analysis (Duarte et al., 1999) and more recently by an extensive and combined gene analysis (Kurtzman and Robnett, 2003). The restriction profile of the 26S rDNA enabled the separation of the Kazachstania exigua (formerly named as Saccharomyces exiguus), a species member of Saccharomycetaceaefamily.
The grouping of Zygosaccharomyces bailii and Z. lentus, in one cluster allowed separating these species which can be very important for quality control purposes (cluster number 4, Figure 2). According to phylogenetic data of the 18S rRNA gene and the ITS region some strains that were previously identified as Z. bailii were reclassified as new species Z. lentus (Steels et al., 1999). This new species also showed some physiological differences when compared to Z. bailii. The remaining studied Zygosaccharomyces species (Z. bisporus and Z. mellis) and Zygotorulaspora florentinus (formerly named as Z. florentinus) presented species-specific restriction profiles.
Hanseniaspora species (anamorph Kloeckera sp.) are common yeast constituents on grapes and often dominate the early stages of wine fermentations (Romano et al., 1993). Growth of these apiculate yeasts may contribute to the final wine quality through production of esters, glycerol and acetoin (Gil et al., 1996). On the other hand, Hanseniaspora sp. may also negatively affect wine fermentations (du Toit and Pretorius, 2000). High levels of this yeast have been found in damaged grapes and might be associated with stuck fermentations (Bisson, 1999). The last unsolved cluster was constituted by H. uvarum and H. guilliermondii which present a very close relatedness (cluster number 5, Figure 2). These two species showed an insignificant D1/D2 sequence divergence which did not exceed 1% (Kurtzman and Robnett, 1998; Cadez et al., 2003), a value that is considered the borderline of species separation (Kurtzman and Robnett, 1998). Indeed, recent results have showed that D. anomala presented a high similarity with these two Hanseniaspora species in restriction profile 5.8S-ITS region, after the digestion with HinfI, HaeIIIand CfoI enzymes (Barata et al., 2008). These authors only achieved the differentiation of H. uvarum from H. guilliermondii and D. anomala by using physiological and biochemical tests. In this work, a separation of Hanseniaspora and Dekkera genus was achieved, highlighting the advantage of using 26S rDNA instead of 5.8 S- ITS region. Nonetheless, for an accurate identification of Hanseniaspora species, sequencing of the ITS regions might be needed (Cadez et al., 2003). In this study, Hanseniaspora occidentalis and H. osmophila presented species-specific restriction profiles.
In order to simplify wine yeast identification using the generated restriction profiles database, cluster analysis was also performed to all possible combinations of three restriction enzymes. The combination of the profiles obtained with the restriction enzymes MseI, HaeIII and CfoI revealed the highest discrimination power. A total of 46 distinct clusters were formed, from which 42 were assigned to a single species (Figure 3). The main difference from the separation achieved with the five restriction enzymes is that S. bayanus, S. pastorianus, T. delbrueckii were grouped together with Z. bailii and Z. lentus. The very close relationship between Zygosaccharomyces, Saccharomyces and Torulaspora genera has already been suggested based on the phylogenetic trees deduced from 18S rDNA (James et al., 1996; 1997) and 26S rDNA (Kurtzman and Robnett, 1998). The closeness between these three genera regarding their response similarity to several physiological tests has also been reported (Esteve-Zarzoso et al., 2003). However, the ApaI enzyme enabled the generation of a distinctive profile for the two Zygosaccharomyces species, therefore allowing their separation from S.bayanus, S. pastorianus and T. delbrueckii (Figure 4). This additional restriction enzyme would be used only if it is necessary to clarify this situation. For example, in wine quality control might be necessary to identify Zygosaccharomyces species which are considered dangerous wine spoilage yeasts as they can produce off-flavors, are osmotolerant, fructophiles, highly-fermentative, tolerant to high ethanol levels and extremely preservative-resistant (Steels et al., 2000; Loureiro and Malfeito-Ferreira, 2003).
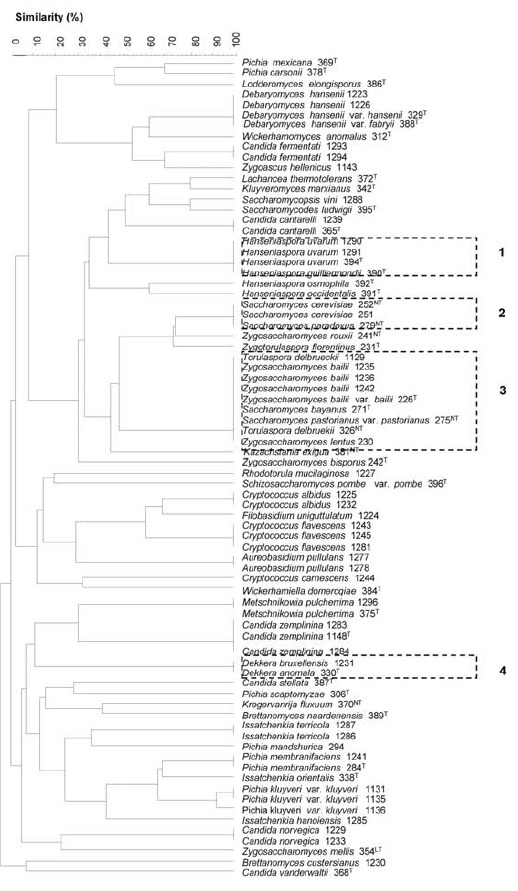
Figure 3 - Dendrogram of restriction profiles presented by the 78 yeast strains by after digestion with MseI, HaeIII and CfoI, generated by using Unweighted Pair Group Method using Arithmetic Average (UPMGA) clustering algorithm (GelCompar version 5.1). The four clusters that could not be solved at species level are shown in dotted lines. The number following each species corresponds to the access number of Colecção de Microrganismos EVN (INRB/INIA Dois Portos); (T), (NT) and (LT) mean type, neotype and lectotype yeasts, respectively.
Dendrograma representando a semelhança entre as 78 estirpes de leveduras com base nos perfis de restrição de MseI, HaeIII and CfoI de uma região do ADNr 26S. Os quatro grupos em que não foi possível a identificação a nível de espécies estão indicados por linhas pontilhadas. O número que segue a espécie de cada estirpe corresponde ao número de entrada na Colecção de Microrganismos EVN (INRB/INIA Dois Portos); (T), (NT) e (LT) significam leveduras tipo, leveduras neotipo e levedura lectótipo, respectivamente.
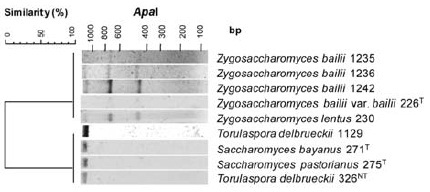
Figure 4 - Dendrogram of restriction profile presented by nine yeast strains after the digestion with ApaI enzyme. The number following each species corresponds to the access number of Colecção de Microrganismos EVN (INRB/INIA Dois Portos); (T) and (NT) means type and neotype yeasts, respectively.
Dendrograma relativo aos perfis de restrição de nove estirpes de leveduras, obtidos após a digestão com a enzima ApaI . O número que segue a espécie de cada estirpe corresponde ao número de entrada na Colecção de Microrganismos EVN (INRB/INIA Dois Portos); (T) e (NT) significam leveduras tipo e neotipo, respectivamente.
CONCLUSIONS
The analysis of the restriction profiles obtained from the PCR amplified NL1-LR6 region of the 26S rDNA allowed the discrimination of 42 species among the 53 yeast species analyzed in this study. The remaining groups comprise closely related species both at taxonomic and wine making levels. The method pointed out in this study represents a fast, less laborious and less expensive technique when compared to sequencing besides it does not require sophisticated equipment. This method is a very useful tool when there is a large number of isolates to be identified. Another practical applicability of the method relies on the capacity to clearly assign the common wine spoilage yeasts D. anomala and D. bruxellensis to one cluster and Z. bailii and Z. lentus to another distinctive cluster. This is an important result in terms of the applicability of the method for quality control purposes. This study allowed the establishment of a restriction profile database based on certified yeast strains that can be used in yeast identification carried out both at research and industrial level.
ACKNOWLEDGEMENTS
The authors thank M. Filomena Alemão for technical assistance. This research was partially supported by the program POCI 2010 (FEDER/FCT, POCTI/AGR/56102/2004).
REFERENCES
Arias C.R., Burns J.K., Friedrich L.M., Goodrich R.M., Parish M.E., 2002. Yeast species associated with orange juice: evaluation of different identification methods. Appl.Environ. Microbiol., 68, 1955-1961.
Baleiras-Couto M.M., Vogels J.T.W.E., Hofstra H., Huis int Veld J.H.J. and van der Vossen, J.M.B.M., 1995. Random amplified polymorphic DNA and restriction enzyme analysis of PCR amplified rDNA in taxonomy: two identification techniques for foodborne yeasts. J. Appl. Bacteriol., 79: 525–535.
Baleiras-Couto M.M., Hartog B.J., Huis in't Veld J.H.J., Hofstra H., van der Vossen J.M.B.M., 1996. Identification of spoilage yeasts in a food-production chain by microsatellite polymerase chain reaction fingerprinting. Food Microbiol., 13, 59-67.
Baleiras-Couto M.M., Reizinho R.G., Duarte F.L., 2005. Partial 26S rDNA restriction analysis as a tool to characterise non-Saccharomyces yeasts present during red wine fermentations. Int. J. Food Microbiol., 102, 49-56.
Barata A., Seborro F., Belloch C., Malfeito-Ferreira M., Loureiro V., 2008. Ascomycetous yeast species recovered from grapes damaged by honeydew and sour rot. J. Appl. Microbiol., 104, 1182-1191.
Bartowsky E.J., Pretorius I.S., 2009. Microbial formation and modification of flavor and off-flavor compounds in Wine. In: Biology of Microorganisms on Grapes, in Must and in Wine. 209-231. König H., Unden G., Frohlich J. (eds), Springer-Verlag, Berlin, Heidelberg.
Bely M., Stoeckle P., Masneuf-Pomarède I., Dubourdieu D., 2008. Impact of mixed Torulaspora delbrueckii-Saccharomyces cerevisiae culture on high-sugar fermentation. Int. J. Food Microbiol. 122, 312-320.
Bisson L.F., 1999. Stuck and sluggish fermentations. Am. J. Enol. Vitic., 50, 107-119.
Bisson L., Joseph, L.C.M.., 2009. Yeasts. In: Biology of microorganisms on grapes, in must and in wines.47-60. König H., Unden G., Fröhlich J. (eds), Springer-Verlag, Berlin-Heidelberg.
Cabrera M.J., Moreno J., Ortega J.M., Medina M., 1988. Formation of ethanol, higher alcohols, esters, and terpenes by five yeast strains in musts from Pedro Ximenez grapes in various degrees of ripeness. Am. J. Enol. Vitic., 39, 283-287.
Cadez, N., Poot, G.A., Raspor, P., and Smith, M.T., 2003. Hanseniaspora meyeri sp. nov., Hanseniaspora clermontiae sp. nov., Hanseniaspora lachancei sp. nov. and Hanseniaspora opuntiae sp. nov., novel apiculate yeast species. Int. J. Syst. Evol. Microbiol., 53:1671-1680.
Ciani M., Ferraro L.1998. Combined use of immobilized Candida stellata cells and Saccharomyces cerevisiae to improve the quality of wines. J. Appl. Microbiol., 85, 247-254.
Ciani M., Maccarelli F., 1998. Oenological properties of non-Saccharomyces yeasts associated with wine-making. World J. Microbiol. Biotech., 14, 199-203.
Ciani M., Beco L., Comitini F., 2006. Fermentation behaviour and metabolic interactions of multistarter wine yeast fermentations. Int. J. Food Microbiol., 108, 239-245.
Clemente-Jimenez J.M., Mingorance-Cazorla L., Martínez-Rodríguez S., Heras-Vázquez F.J.L., Rodríguez-Vico F., 2004. Molecular characterization and oenological properties of wine yeasts isolated during spontaneous fermentation of six varieties of grape must. Food Microbiol., 21, 149-155.
Csoma H., Sipiczki M., 2008. Taxonomic reclassification of Candida stellata strains reveals frequent occurrence of Candida zemplinina in wine fermentation. FEMS Yeast Res. 8, 328-336.
Dlauchy D., Tornai-Lehoczki J., Péter G., 1999. Restriction enzyme analysis of PCR amplified rDNA as a taxonomic tool in yeast identification. Syst. Appl. Microbiol. 22, 445-453.
Domizio P., Lencioni L., Ciani M., Di Blasi S., Pontremolesi C., Sabatelli M.P., 2007. Spontaneous and inoculated yeast populations dynamics and their effect on organoleptic characters of Vinsanto wine under different process conditions. Int. J. Food Microbiol., 115, 281-289.
Duarte F.L., Pais C., Spencer-Martins I., Leão C., 1999. Distinctive electrophoretic isoenzyme profiles in Saccharomyces sensu stricto. Int. J. Syst. Bacteriol., 49, 1907-1913.
Duarte F.L., Pais C., Spencer-Martins I., Leão C., 2004. Isoenzyme patterns: a valuable molecular tool for the differentiation of zygosaccharomyces species and detection of misidentified isolates. Syst.Appl.Microbiol., 27, 436-442.
Du Toit M., Pretorius I.S., 2000. Microbial spoilage and preservation of wine: using weapons for nature's own arsenal – A review. South Afr. J. Enol.Vitic., 21, 74-96.
Esteve-Zarzoso B., Belloch C., Uruburu F., Querol A., 1999. Identification of yeasts by RFLP analysis of the 5.8S rRNA gene and the two ribosomal internal transcribed spacers. Int. J. Syst. Bacteriol., 49, 329-337.
Esteve-Zarzoso B., Zorman T., Belloch C., Querol A., 2003. Molecular characterisation of the species of the genus Zygosaccharomyces. Syst. Appl. Microbiol., 26, 404-411.
Fernández-Espinar M.T., Martorell P., Llanos R., Querol A., 2006. Molecular methods to identify and characterize yeasts in foods and beverages. In: Yeasts in Food and Beverages (Q. Amparo, and G.H. Fleet, eds), Heidelberg: Springer-Verlag, pp 55-82.
Ferreira A.M., Clímaco M.C., Faia A.M., 2001. The role of non-Saccharomyces species in releasing glycosidic bound fraction of grape aroma components - a preliminary study. J. Appl. Microbiol., 91, 67-71.
Ferreira N., Belloch C., Querol A., Manzanares P., Vallez S., Santos A.,2010. Yeast microflora isolated from Brazilian cassava roots: taxonomical classification based on molecular identification. Curr. Microbiol., 60, 287-293.
Fleet G.H., 2003. Yeast interactions and wine flavour. Int. J. Food Microbiol., 86, 11-22.
Fleet G.H., Heard G.M., 1993. Yeasts: growth during fermentation. In: Wine Microbiology and Biotechnology, 27-54. Fleet G.H. (ed), Harwood Academic, Chur, Switzerland.
Gil J., Mateo J., Jiménez M., Pastor A., Huerta T., 1996. Aroma compounds in wine as influenced by apiculate yeasts. J. Food Sci., 61, 1247-1250.
González S., Barrio E., Querol A., 2007. Molecular identification and characterization of wine yeasts isolated from Tenerife (Canary Island, Spain). J. Appl. Microbiol., 102 1018-1025.
Guillamón J.M., Sabaté J., Barrio E., Cano J., Querol A., 1998. Rapid identification of wine yeast species based on RFLP analysis of the ribosomal internal transcribed spacer (ITS) region. Arch. Microbiol., 169, 387-392.
Hernán-Gómez S., Espinosa J.C., Ubeda J.F., 2000. Characterization of wine yeasts by temperature gradient gel electrophoresis (TGGE). FEMS Microbiol. Lett., 193, 45-50.
Hierro N., González Á., Mas A., Guillamón J.M., 2006. Diversity and evolution of non-Saccharomyces yeast populations during wine fermentation: effect of grape ripeness and cold maceration. FEMS Yeast Res., 6, 102-111.
Hierro N., Esteve-Zarzoso B., Mas A., Guillamón J.M., 2007. Monitoring of Saccharomyces and Hanseniaspora populations during alcoholic fermentation by real-time quantitative PCR. FEMS Yeast Res., 7, 1340-1349.
James S.A., Collins M.D., Roberts I.N., 1996. Use of an rRNA Internal Transcribed Spacer region to distinguish phylogenetically closely related species of the genera Zygosaccharomyces and Torulaspora. Int. J. Syst. Bacteriol., 46, 189-194.
James S.A., Cai J., Roberts I.N., Collins M.D., 1997. A Phylogenetic Analysis of the Genus Saccharomyces Based on 18S rRNA Gene Sequences: Description of Saccharomyces kunashirensis sp. nov. and Saccharomyces martiniae sp. nov. Int. J. Syst. Bacteriol., 47, 453-460.
Kurtzman C.P., Robnett C.J., 1994. Synonymy of the yeast genera Wingea and Debaryomyces. Antonie van Leeuwenhoek, 66, 337-342.
Kurtzman C.P., Fell J.W., 1998. The yeasts: a taxonomic study. 1076 p. Elsevier Science, Amsterdam, The Netherlands.
Kurtzman C.P., Robnett C.J., 1998. Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie van Leeuwenhoek, 73, 331-371.
Kurtzman C.P., Robnett C.J., 2003. Phylogenetic relationships among yeasts of the 'Saccharomyces complex' determined from multigene sequence analyses. FEMS Yeast Res., 3, 417-432.
Kurtzman C.P., Pikur J., 2005. Taxonomy and phylogenetic diversity among the yeasts. In: Topics in Current Genetics. 29-46. Sunnerhagen P., Pikur J. (eds), Springer-Verlag, Berlin-Heidelberg.
Longo E., Cansado J., Agrelo D., Villa T.G., 1991., Effect of climatic conditions on yeast diversity in grape musts from northwest spain. Am.J. Enol. Viticult., 42, 141-144.
Loureiro V., Malfeito-Ferreira M., 2003. Spoilage yeasts in the wine industry. Int. J. Food Microbiol., 86, 23-50.
Mardis E.R., 2008. The impact of next-generation sequencing technology on genetics. Trends Genet., 24,133–141.
Martini A., 1992. Biodiversity and conservation of yeasts. Biodiv. & Conserv.,1, 324-333. [ Links ]
Mills D.A., Johannsen E.A., Cocolin L., 2002. Yeast diversity and persistence in Botrytis-affected wine fermentations. Appl. Environ. Microbiol., 68, 4884-4893.
Naumova E.S., Bulat S.A., Mironenko N.V., Naumov G.I., 2003. Differentiation of six sibling species in the Saccharomyces sensu stricto complex by multilocus enzyme electrophoresis and UP-PCR analysis. Antonie van Leeuwenhoek, 83, 155-166.
Nisiotou A.A., Spiropoulos A.E., Nychas G-J.E., 2007. Yeast community structures and dynamics in healthy and Botrytis-affected grape must fermentations. Appl. Environ. Microbiol., 73: 6705-6713.
Pretorius I.S., 2000. Tailoring wine yeast for the new millennium: novel approaches to the ancient art of winemaking. Yeast, 16, 675-729.
Renouf V., Lonvaud-Funel A., 2007. Development of an enrichment medium to detect Dekkera/Brettanomyces bruxellensis, an spoilage wine yeast, on the surface of grape berries. Microbiol. Res., 162, 154-167. [ Links ]
Rodríguez M.E., Infante J.J., Molina M., Domínguez M., Rebordinos L., Cantoral J.M., 2010. Genomic characterization and selection of wine yeast to conduct industrial fermentations of a white wine produced in a SW Spain winery. J. Appl. Microbiol., 108, 1292-1302.
Romano P., Suzzi G., Comi G., Zironi R., Maifreni M., 1997. Glycerol and other fermentation products of apiculate wine yeasts. J. Appl. Microbiol., 82, 615-618.
Romano P., Suzzi G., Zironi R., Comi G., 1993. Biometric study of acetoin production in Hanseniaspora guilliermondii and Kloeckera apiculata. Appl. Environ. Microbiol., 59, 1838-1841.
Romano P., Fiore C., Paraggio M., Caruso M., Capece A., 2003. Function of yeast species and strains in wine flavour. Int. J. Food Microbiol., 86,169-180.
Romano P., Capece A., Serafino V., Romaniello R., Poeta C., 2008. Biodiversity of wild strains of Saccharomyces cerevisiae as tool to complement and optimize wine quality. World J. Microbiol. Biotech., 24,1797-1802.
Salinas F., Garrido D., Ganga A., Veliz G., Martínez C., 2009. Taqman real-time PCR for the detection and enumeration of Saccharomyces cerevisiae in wine. Food Microbiol., 26, 328-332.
Sampaio J.P., Gadanho M., Santos S., Duarte F.L., Pais C., Fonseca A. Fell J.W., 2001. Polyphasic taxonomy of the basidiomycetous yeast genus Rhodosporidium: Rhodosporidium kratochvilovae and related anamorphic species. Inter. J. Syst. Evol. Microbiol., 51, 687-697.
Santos A., San Mauro M., Bravo E., Marquina D., 2009. PMKT2, a new killer toxin from Pichia membranifaciens, and its promising biotechnological properties for control of the spoilage yeast Brettanomyces bruxellensis. Microbiol., 155, 624-634. [ Links ]
Schütz M., Gafner J., 1993. Analysis of yeast diversity during spontaneous and induced alcoholic fermentations. J. Appl. Bacteriol.75, 551-558.
Smith M.T., Yamazaki M., Poot G.A., 1990. Dekkera, Brettanomyces and Eeniella: Electrophoretic comparison of enzymes and DNA–DNA homology. Yeast, 6, 299-310.
Steels H., Bond C.J., Collins M.D., Roberts I.N., Stratford M., James S.A., 1999. Zygosaccharomyces lentus sp. nov., a new member of the yeast genus Zygosaccharomyces Barker. Int. J. Syst. Bacteriol., 49, 319-327.
Steels H., James S.A., Roberts I.N., Stratford M., 2000. Sorbic acid resistance: the inoculum effect. Yeast, 16, 1173-1183.
Tessonnière H., Vidal S., Barnavon L., Alexandre H., Remize F., 2009. Design and performance testing of a real-time PCR assay for sensitive and reliable direct quantification of Brettanomyces in wine. Int. J. Food Microbiol., 129, 237-243.
Torija M.J., Rozès N., Poblet M., Guillamón J., Mas A., 2001. Yeast population dynamics in spontaneous fermentations: Comparison between two different wine-producing areas over a period of three years. Antonie van Leeuwenhoek, 79, 345-352.
Török T., Rockhold D., and King Jr A.D., 1993. Use of electrophoretic karyotyping and DNA-DNA hybridization in yeast identification. Int. J. Food Microbiol., 19, 63-80.
White T.J., Bruns T., Lee S., Taylor J.W., 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: PCR protocols: a guide to methods and applications. Innis M.A., Gelfand D.H., Sninsky J.J., White T.J. (eds), Academic Press, New York, pp 315-322.
Zott K., Miot-Sertier C., Claisse O., Lonvaud-Funel A., Masneuf-Pomarede I., 2008. Dynamics and diversity of non-Saccharomyces yeasts during the early stages in winemaking. Int. J. Food Microbiol.,125, 197-203.
Zott K., Claisse O., Lucas P., Coulon J., Lonvaud-Funel A., Masneuf-Pomarede I., 2010. Characterization of the yeast ecosystem in grape must and wine using real-time PCR. Food Microbiol., 27, 559-567.
*Corresponding author: geni.zanol@inrb.pt
(Manuscrito recebido em 30.11.10. Aceite para publicação em 23.12.10)